

Selection of awardees is handled directly by the organizer who will notify all eligible participants.
#ICLIP EXPERIMENTS PROTOCOLS RNA IP REGISTRATION#
Applicants do not need to apply separately for travel grants for this event but should indicate on the registration form if they wish to be considered for a travel grant. Travel grantsĪ limited number of travel grants are available for eligible participants. All participants will present their poster. You will be provided with velcro tabs and pins for use in adhering your poster to the board. Materials should be printed on thin poster paper or cardboard, anything heavier will not stay in position. Each participant will be provided with a A0 poster board on which to display the poster. The course includes several poster sessions in which participants will explain their research projects.
#ICLIP EXPERIMENTS PROTOCOLS RNA IP FULL#
To see full programme of the course click HERE. Stefanie Ebersberger, Buchmann Institute for Molecular Life Sciences (BMLS), Goethe University Frankfurt, Germany.Anke Busch, Institute of Molecular Biology (IMB), Germany.Maria do Carmo-Fonseca, Faculdade de Medicina da Universidade de Lisboa, Portugal.Matthias Hentze, European Molecular Biology Laboratory, Germany.Kathi Zarnack, Buchmann Instituite for Molecular Life Science (BMLS), Goethe University Frankfurt, Germany.Christopher Sibley, Imperial College London, UK.Michaela Müller-McNicoll, Goethe University Frankfurt, Germany.Julian König, Institute of Molecular Biology (IMB), Germany.
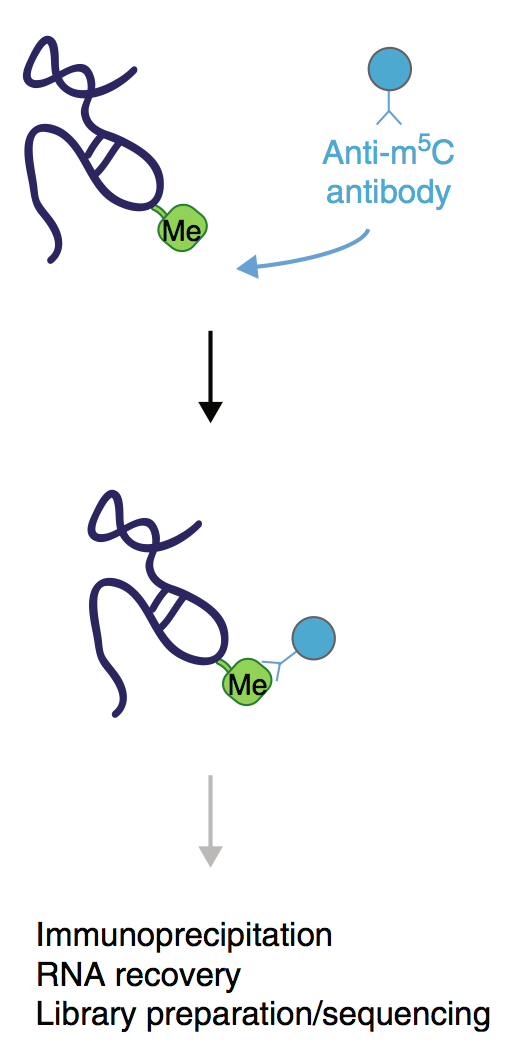
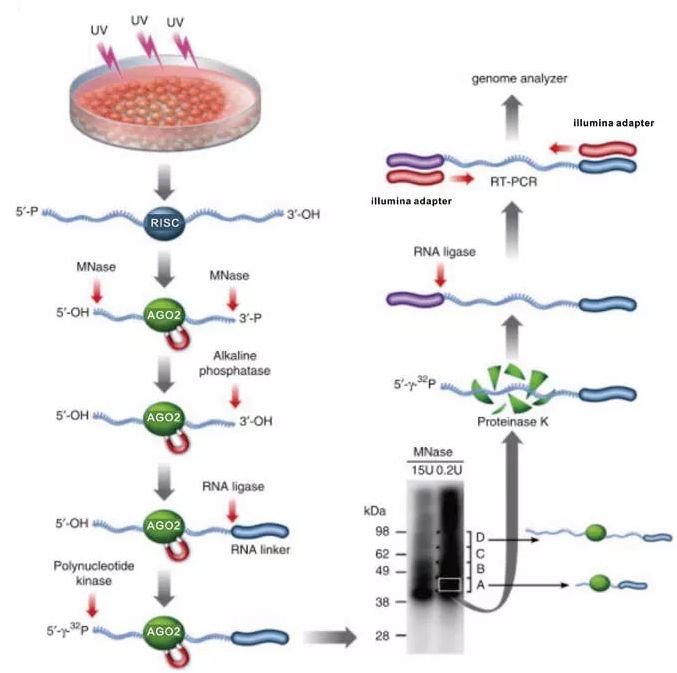
The hands-on sessions in the laboratory will be complemented by presentations by established scientists that will share their experience on using iCLIP to answer different biological questions. We will further discuss the bioinformatics strategies for analysis of iCLIP data. We will guide the participants through the complete protocol, placing special emphasis on critical optimisation steps and controls. The major objective of this EMBO Practical Course is to disseminate the knowledge and proficiency in this technically challenging ribonomics method.
Individual-nucleotide resolution UV crosslinking and immunoprecipitation (iCLIP) has established itself as a leading method to map the RNA interaction sites of a single RBP-of-interest with high resolution and in a quantitative manner. Understanding these interactions in a genome-wide manner is crucial for gaining a complete picture of an RBP’s specificity and function. In turn, RBPs can interact with hundreds to thousands of cellular RNAs by recognizing certain sequence motifs or structural elements. Throughout its life cycle, each RNA will interact dynamically with numerous RNA-binding proteins (RBPs) that determine how the RNA is processed, where it is localized, whether it is translated and finally how it is degraded.


 0 kommentar(er)
0 kommentar(er)
